Overview of IMP3 steps¶
The IMP3 workflow consists of six modules: Preprocessing of reads , Assembly, Analysis of contigs including functional annotation, Binning of contigs, determination of Taxonomy at various levels, and Summary including visualization. Typically, IMP3 will use raw sequencing reads as input and run through the whole workflow.
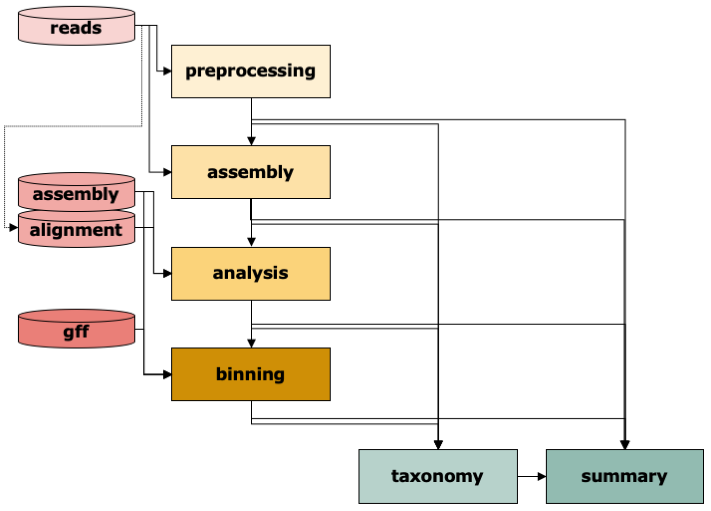
Given the appropriate input, most steps can be run independently. For instance, if the user has
already processed reads and wants to assemble and annotate them. The user would then select only assembly and analysis from the steps (and potentially summary). The user can also supply directly a given assembly to annotate and bin it.
Or an already annotated assembly just for binning etc. Just enter the appropriate input data and define the steps in the config file and IMP3 will take care of it.
To get an impression of all Snakemake steps of a run with metaG and metaT reads as input
using hybrid assembly, download this file.